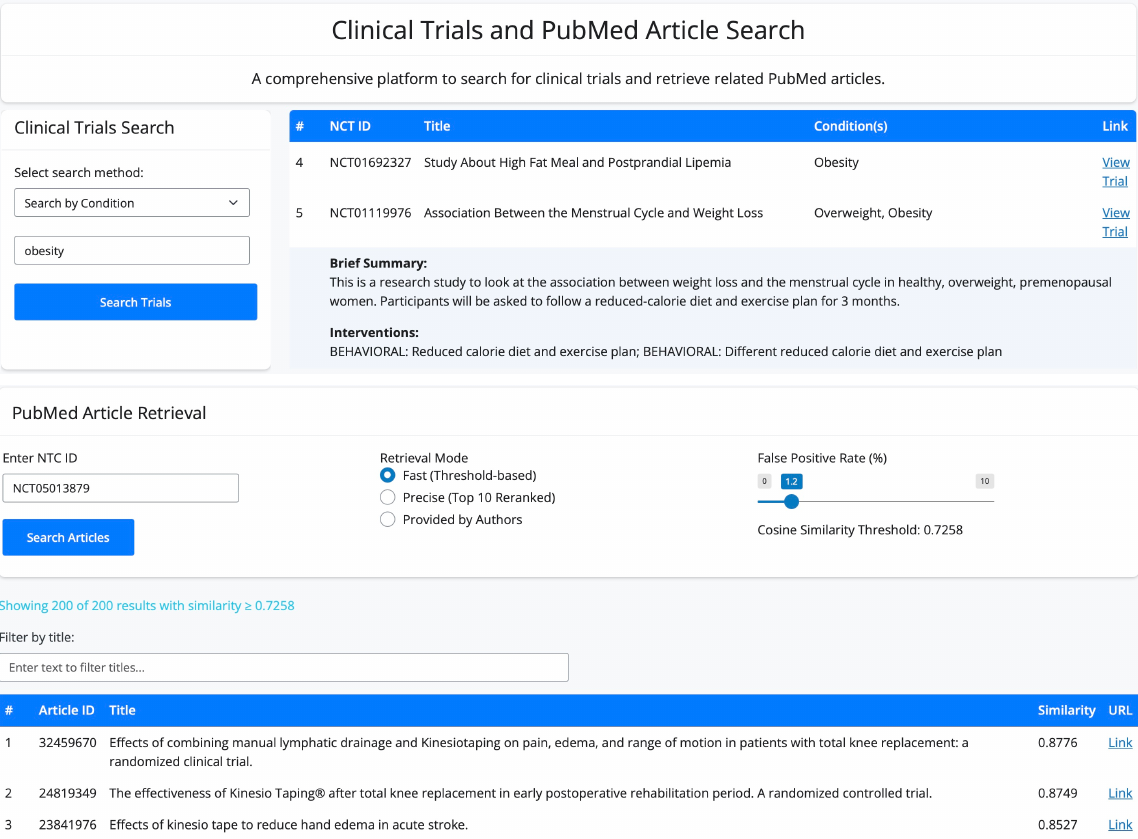
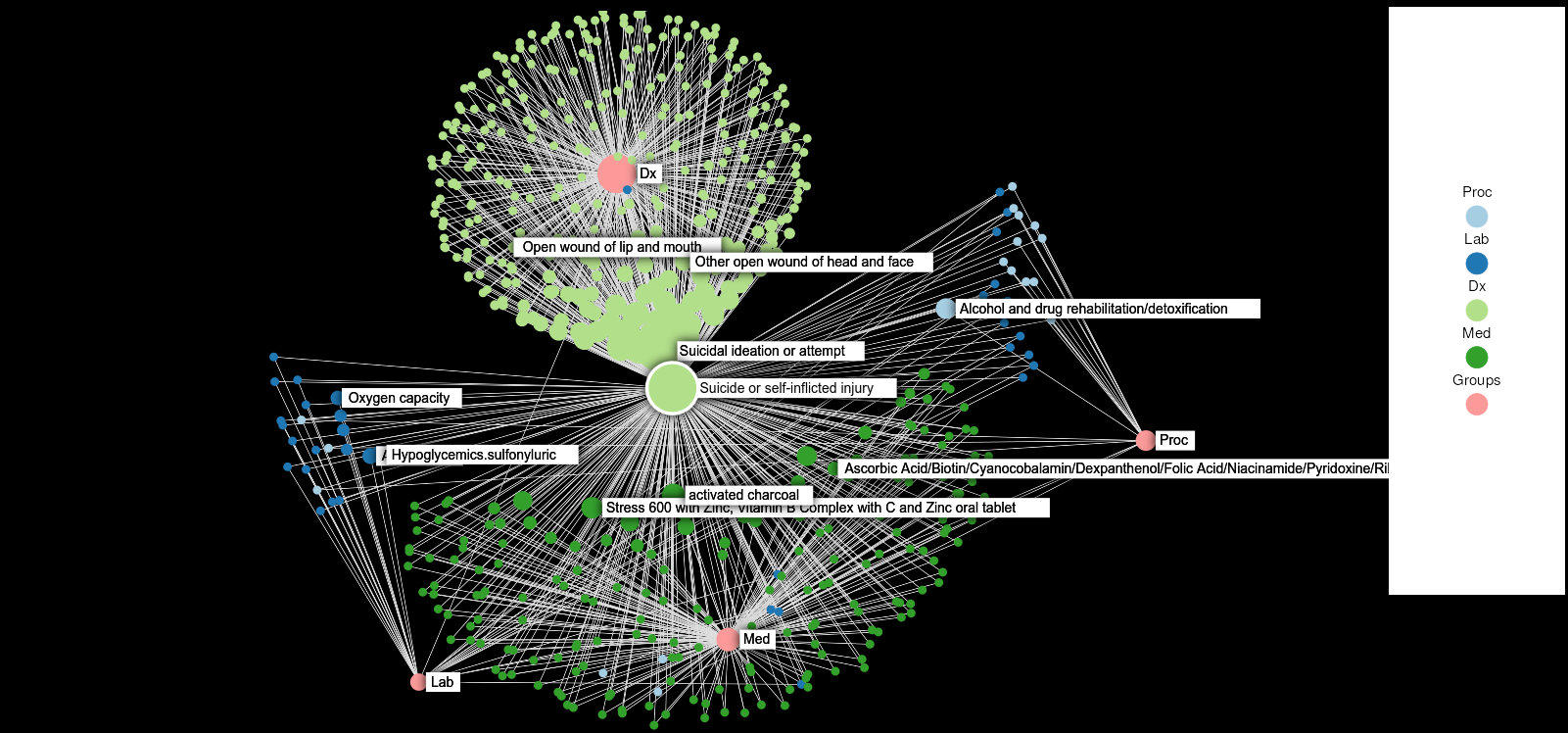
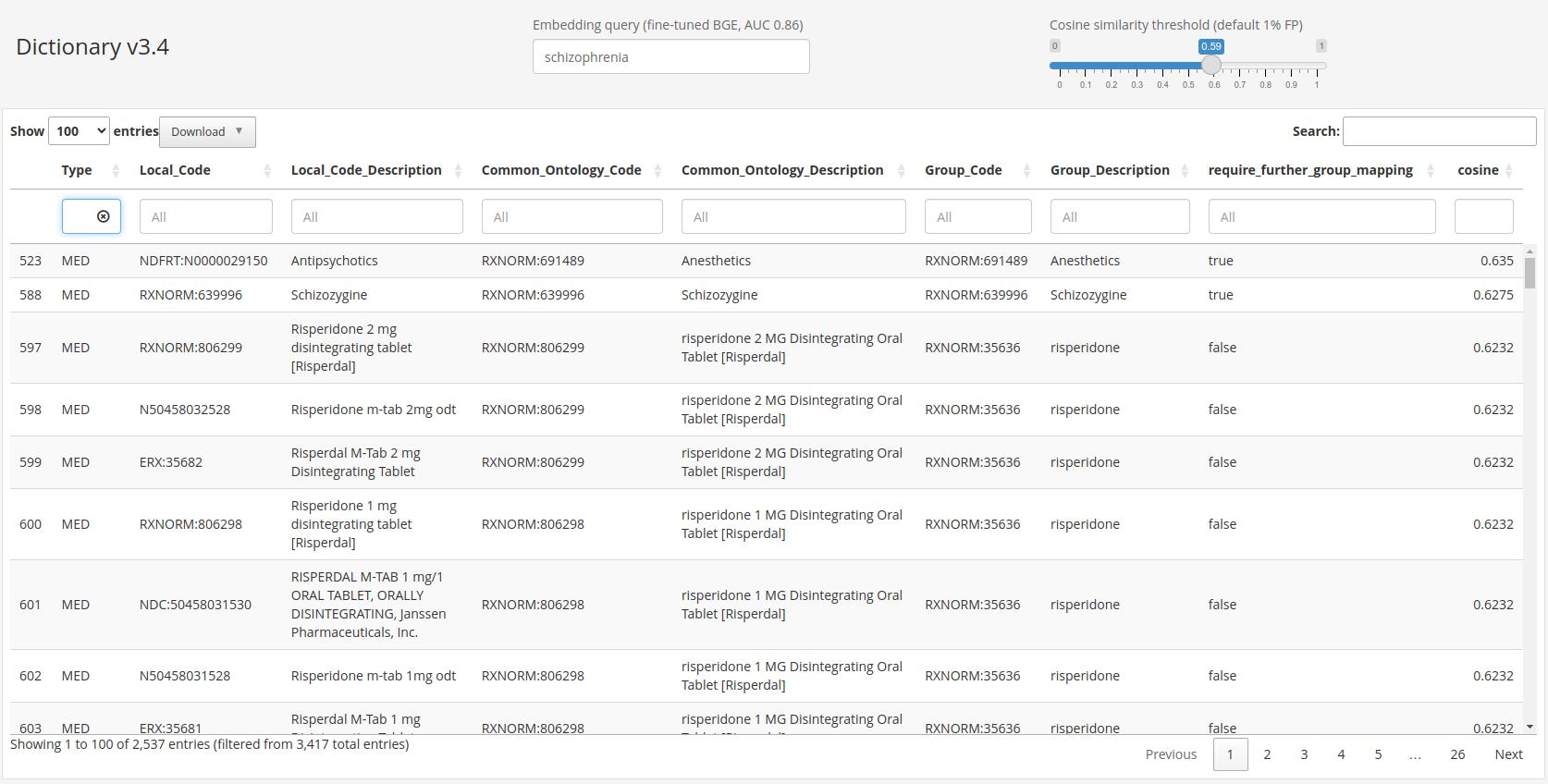
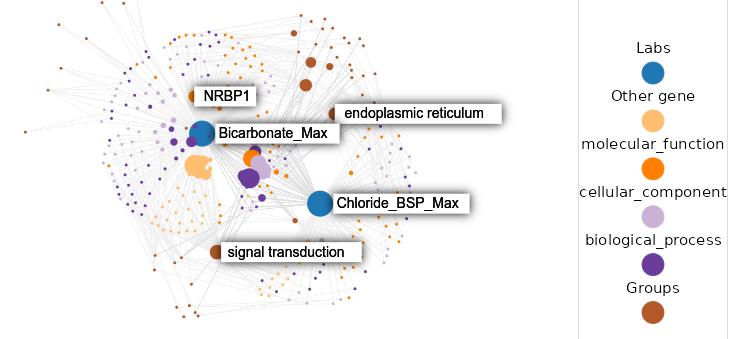
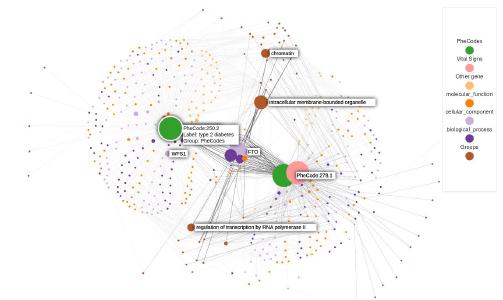
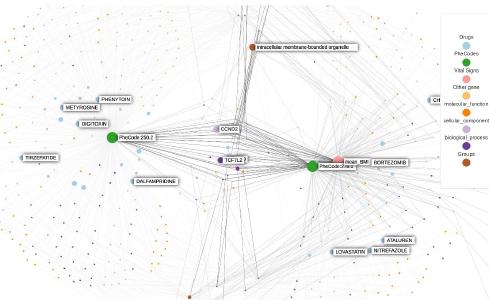
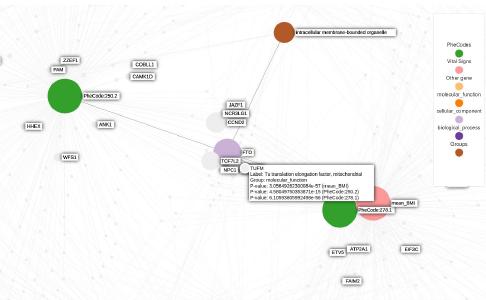
Knowledge graphs enable to efficiently visualize and gain insights into large-scale data analysis results, as p-values from multiple studies or embedding data matrices. The usual workflow is a user providing a data frame of association studies results and specifying target nodes, e.g. phenotypes, to visualize. The knowledge graph then shows all the features which are significantly associated with the phenotype, with the edges being proportional to the association scores. As the user adds several target nodes and grouping information about the nodes such as biological pathways, the construction of such graphs soon becomes complex. The ‘kgraph’ package aims to enable users to easily build such knowledge graphs, and provides two main features: first, to enable building a knowledge graph based on a data frame of concepts relationships, be it p-values or cosine similarities; second, to enable determining an appropriate cut-off on cosine similarities from a complete embedding matrix, to enable the building of a knowledge graph directly from an embedding matrix. The ‘kgraph’ package provides several display, layout and cut-off options, and has already proven useful to researchers to enable them to visualize large sets of p-value associations with various phenotypes, and to quickly be able to visualize embedding results. Two example datasets are provided to demonstrate these behaviors, and several live ‘shiny’ applications are hosted by the CELEHS laboratory and Parse Health, as the KESER Mental Health application https://keser-mental-health.parse-health.org/ based on Hong C. (2021) doi:10.1038/s41746-021-00519-z.
Continue reading