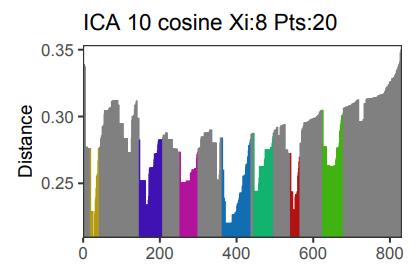
The OPTICS k-Xi density-based clustering pipeline now integrates ensemble metrics which enable to increase the stability of the clusters and the reproducibility of the results on evaluation datasets. Up to now, OPTICS k-Xi computed several distance-based metrics but did not implement any way of merging together their results, it was up to the user to select the most relevant metric or to merge them together. Recently, we applied the pipeline on natural language processing embeddings and the choice of the relevant metric revealed complex and the clusters were unstable. To alleviate this, we implemented ensemble metrics and models in the OPTICS k-Xi pipeline. Three other improvements have also been implemented: the selection of the cosine distance; a parameter to set a maximum cluster size; and a parameter to select a minimum number of clusters. This vignette showcases and introduce the new developments of the opticskxi package, with a particular focus on ensemble metrics and models.
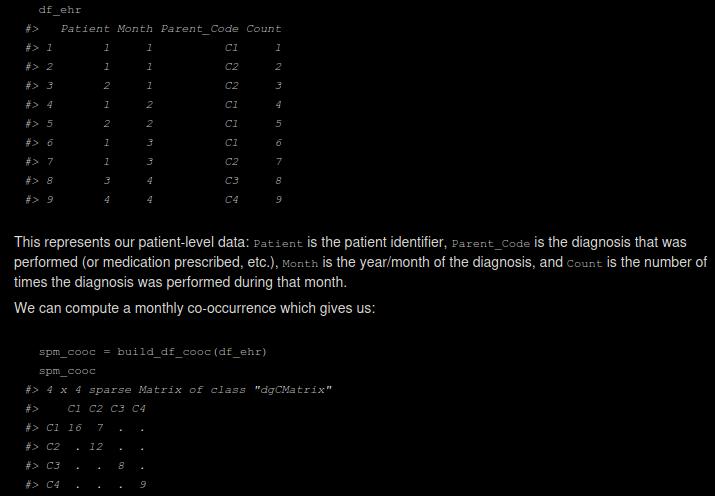
The nlpembeds package provides efficient methods to compute co-occurrence matrices, pointwise mutual information (PMI) and singular value decomposition (SVD) embeddings. In the biomedical and clinical setting, one challenge is the huge size of databases of electronic health records (EHRs), e.g. when analyzing data of millions of patients over tens of years. To address this, this package provides functions to efficiently compute monthly co-occurrence matrices, which is the computational bottleneck of the analysis, by using the RcppAlgos package and sparse matrices. Furthermore, the functions can be called on SQL databases, enabling the computation of co-occurrence matrices of tens of gigabytes of data, representing millions of patients over tens of years. PMI-SVD embeddings are extensively used, e.g. in Hong C. (2021).
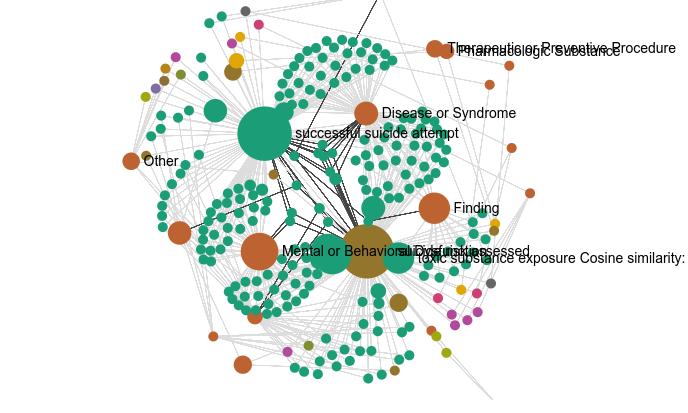
Knowledge graphs enable to organize vast amounts of knowledge and has been used extensively on internet to help websites share data and integrate one another. In the domain of research and knowledge discovery, they enable to organise results and provide specific views of results by showing subsets of interest.
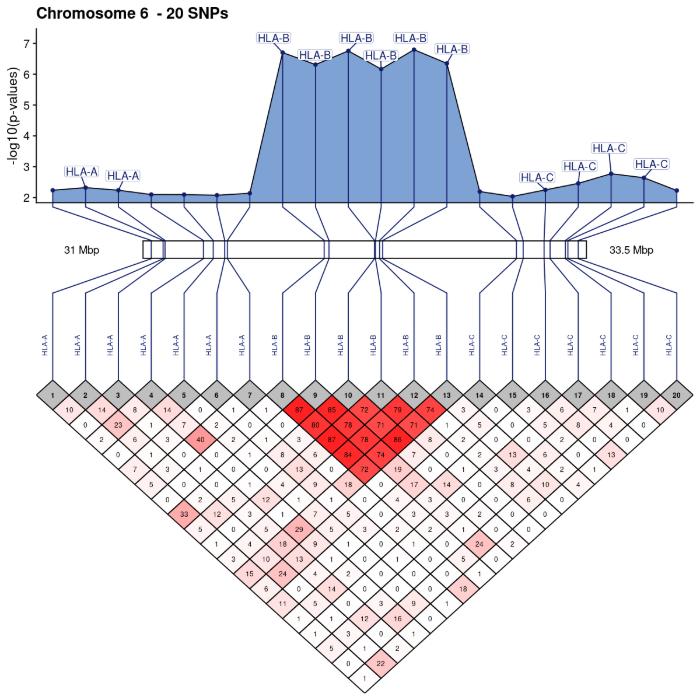
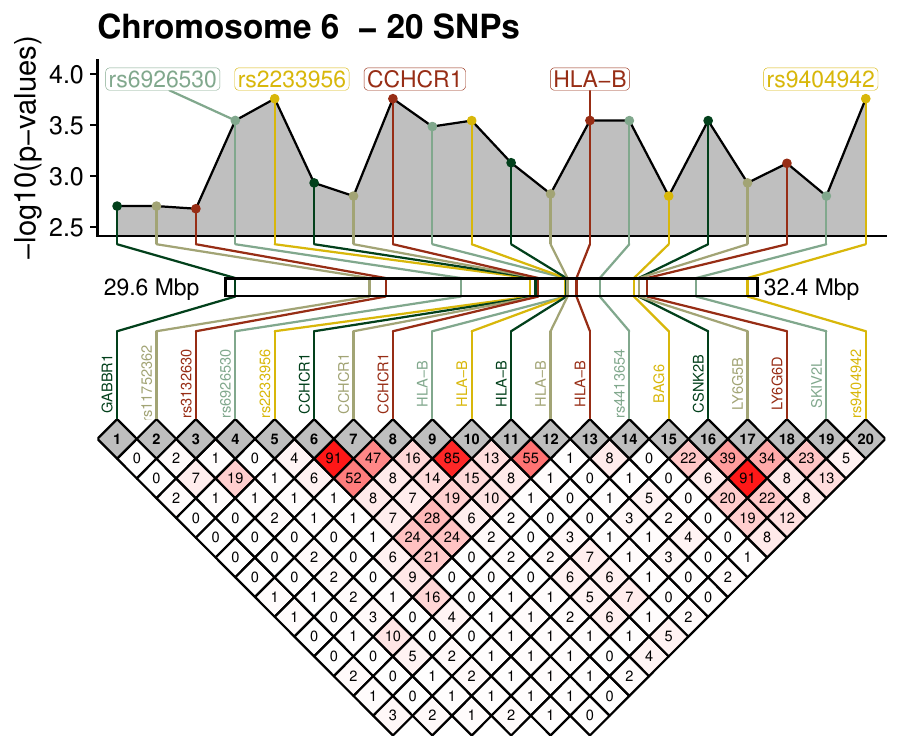
SNPLinkage is modular enough to use directly dataframes of correlation matrices and chromosomic positions specified by the user, e.g. for visualizing RNASeq data. The user can compute a correlation matrix or any kind of pair-wise similarity matrix independently and then use SNPLinkage to build and arrange easily customizable ggplot2 objects.
Single nucleotide polymorphisms are the most common genetic variations in humans and genome-wide microarrays measure up to several millions. Physically close polymorphisms often exhibit correlation structures and are said to be in linkage disequilibrium when they are more correlated than randomly expected. The snplinkage package provides linkage disequilibrium visualizations by displaying correlation matrices annotated with chromosomic positions and gene names. Two types of displays are provided to focus on small or large regions, and both can be extended to combine associations results or investigate feature selection methods. This article introduces the package and illustrates the variety of correlation structures found genome-wide by focusing on 3 regions associated with autoimmune diseases: the chromosome 1 region 1p13.2 (111-115 Mbp), the chromosome 8 region 8p23.1 (7-12 Mbp) and the chromosome 6 region 6p21.3 (29-35 Mbp).
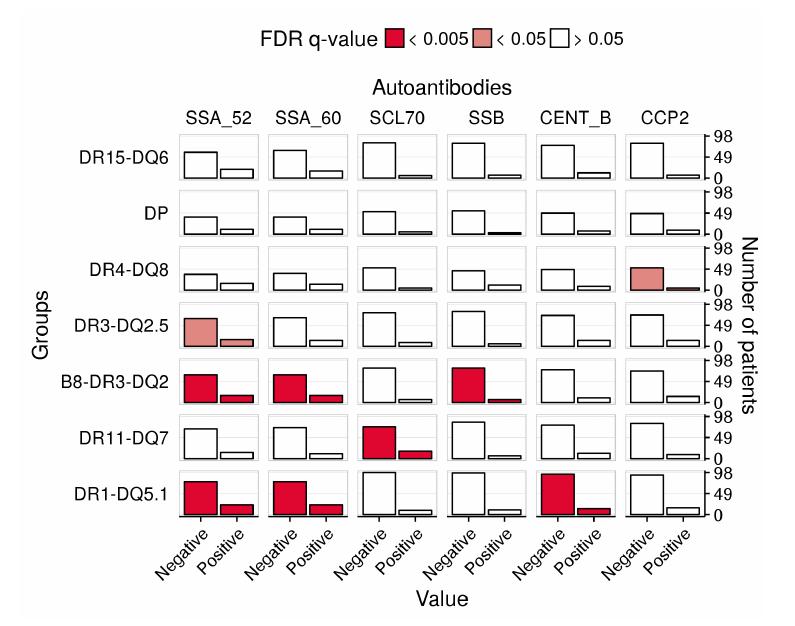
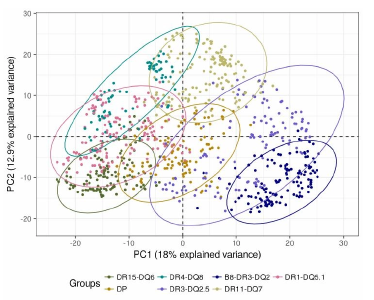
Systemic autoimmune diseases are considered to share genetic susceptibility markers and clinicians expect treatments could benefit from a molecular-based reclassification. In that objective, more than 1,000 patients were recruited by the PRECISESADS project to measure their genotypes and their proteins concentrations in blood.
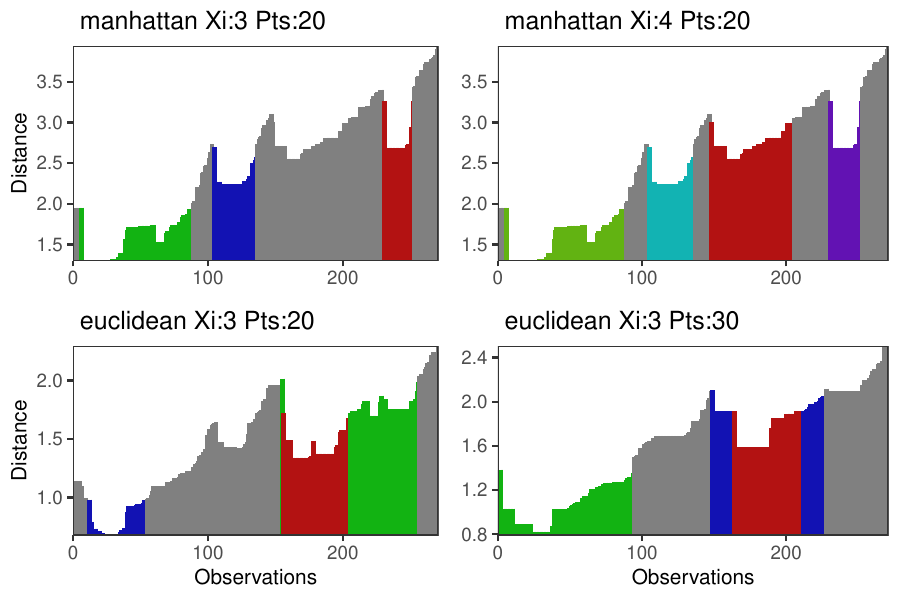
Density-based clustering methods are well adapted to the clustering of high-dimensional data and enable the discovery of core groups of various shapes despite large amounts of noise. The opticskxi R package provides a novel density-based cluster extraction method, OPTICS k-Xi, and a framework to compare k-Xi models using distance-based metrics to investigate datasets with unknown number of clusters. This article ␜rst introduces density-based algorithms with simulated datasets, then presents and evaluates the k-Xi cluster extraction method. Finally, the models comparison framework is described and experimented on 2 genetic datasets to identify groups and their discriminating features. The k-Xi algorithm is a novel OPTICS cluster extraction method that speci␜es directly the number of clusters and does not require ␜ne-tuning of the steepness parameter as the OPTICS Xi method. Combined with a framework that compares models with varying parameters, the OPTICS k-Xi method can identify groups in noisy datasets with unknown number of clusters.
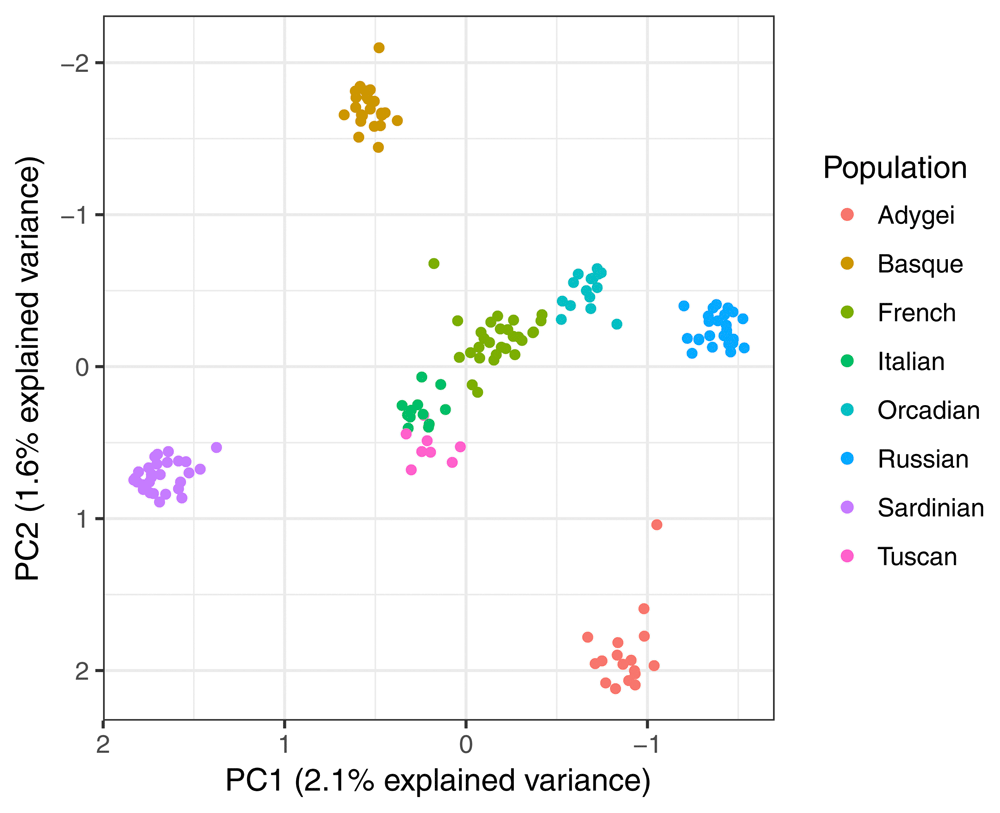
Background. In 2008, several principal component analyses (PCAs) applied on 660,918 single-nucleotide polymorphisms (SNPs) from 938 individuals from 51 worldwide populations of the Human Genome Diversity Panel were published by Li et al. PCAs were applied on subsets of individuals sharing a common geographic origin and showed that in several geographic regions, genome-wide variations of SNPs grouped individuals by populations in the two first principal components. In this study, we replicated the PCAs applied on two geographic subsets, first on individuals from Europe and second on individuals from the Middle East & North Africa. Methods. Quality control, feature selection, and PCA were applied on each geographic subset. The results were displayed on the two first principal components and compared to the original figures. Results. The replicated figures were found to match closely to the original figures. Conclusions. Therefore, the main results were replicated and can be independently reproduced by using publicly available data, source code, and computing environment.
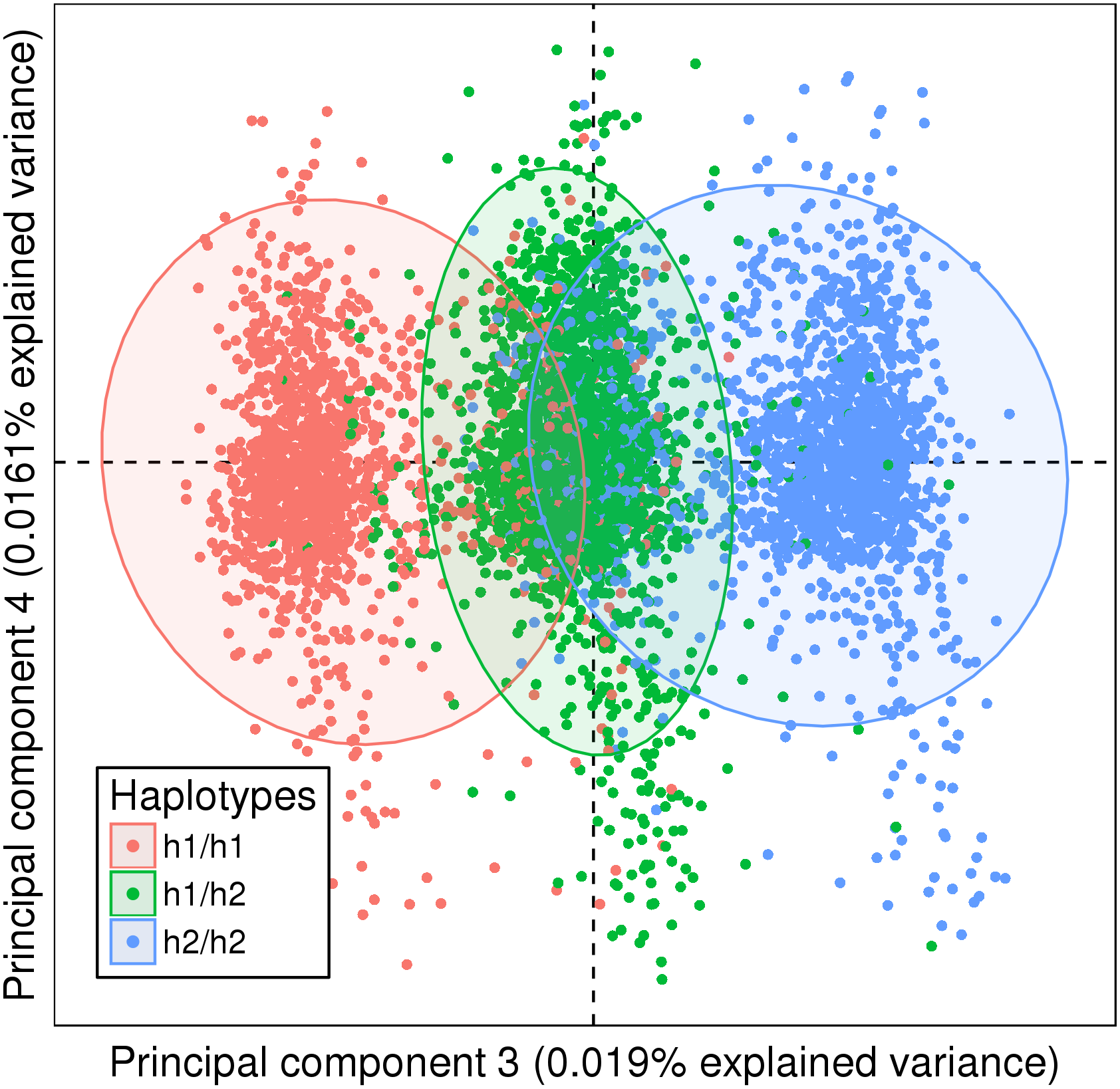
Systemic Autoimmune Diseases, a group of chronic inflammatory conditions, have variable symptoms and difficult diagnosis. In order to reclassify them based on genetic markers rather than clinical criteria, we performed clustering of Single Nucleotide Polymorphisms. However naive approaches tend to group patients primarily by their geographic origin. To reduce this “ancestry signal”, we developed SNPClust, a method to select large sources of ancestry-independent genetic variations from all variations detected by Principal Component Analysis. Applied to a Systemic Lupus Erythematosus case control dataset, SNPClust successfully reduced the ancestry signal. Results were compared with association studies between the cases and controls without or with reference population stratification correction methods. SNPClust amplified the disease discriminating signal and the ratio of significant associations outside the HLA locus was greater compared to population stratification correction methods. SNPClust will enable the use of ancestry-independent genetic information in the reclassification of Systemic Autoimmune Diseases. SNPClust is available as an R package and demonstrated on the public Human Genome Diversity Project dataset at https://github.com/ThomasChln/snpclust.