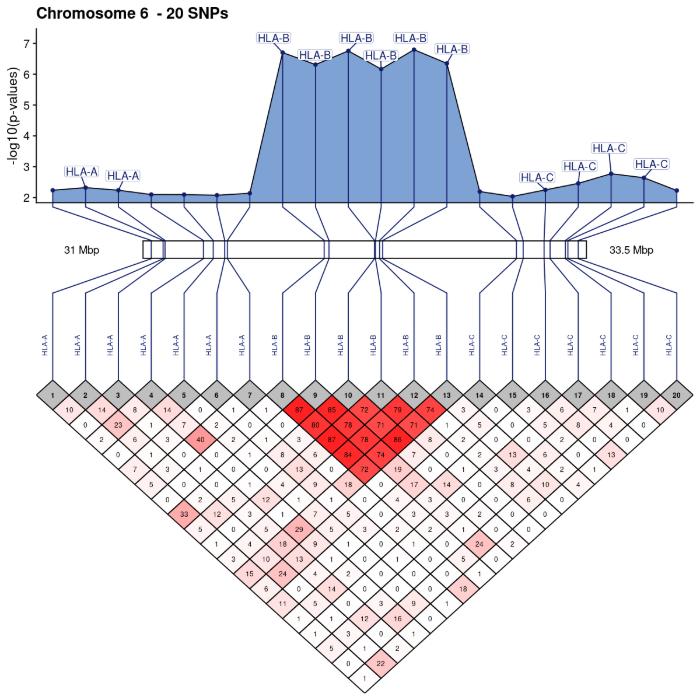
SNPLinkage is modular enough to use directly dataframes of correlation matrices and chromosomic positions specified by the user, e.g. for visualizing RNASeq data. The user can compute a correlation matrix or any kind of pair-wise similarity matrix independently and then use SNPLinkage to build and arrange easily customizable ggplot2 objects.
Links
© All rights reserved, Thomas Charlon, 2025.
Template by Bootstrapious. Ported to Hugo by DevCows.

